15.3 Types of Mutation
Mutations can be classified in a number of different ways. We will first consider large-scale mutations that affect overall chromosomal structure, and then small-scale mutations that change only one or a few nucleotides.
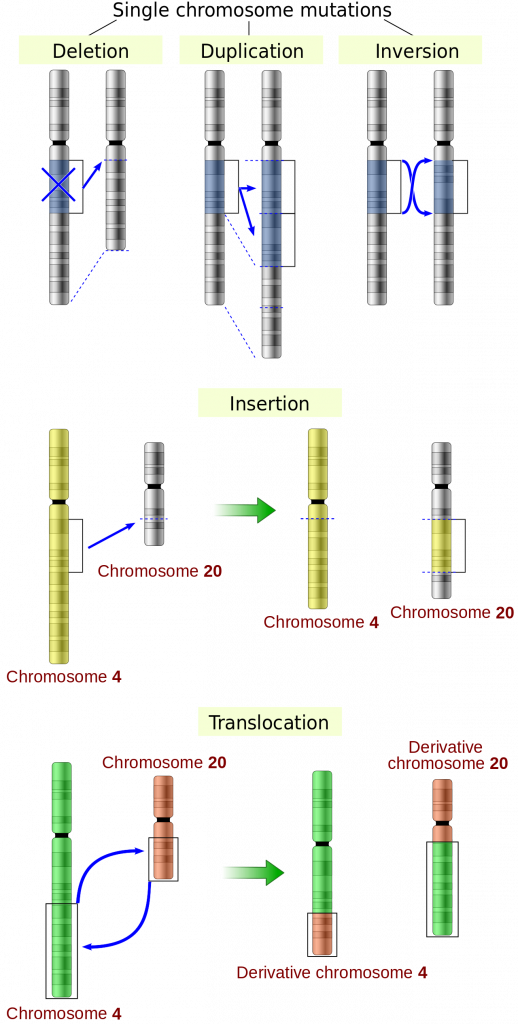
Large-scale mutations include deletions, duplications, or inversions of a single chromosome. Deletions can be deleterious because they may result in the loss of genes. It is important for us to recognize that an average human chromosome contains about 1000 genes. Thus a deletion may remove hundreds of genes. In contrast, duplications can result in extra copies of genes. This can be deleterious at the individual level, if the organism is sensitive to gene dosage. However, gene duplications are extremely important in macroevolution. If a population has two copies of a particular gene, only one needs to be maintained to provide the original function, while the other is free to vary and perhaps take on a new function. Inversions, in which a segment of DNA is reversed in orientation, can be deleterious due to rearrangement of genetic information at the break points.
Large-scale mutations can also involve more than one chromosome. For instance, inversionstranslocation occurs when two chromosomes exchange segments of DNA. This can bring pieces of DNA together, potentially causing fusion genes, which are genes that are formed from two genes that were previously separate. Fusion genes have been identified in many types of cancer.
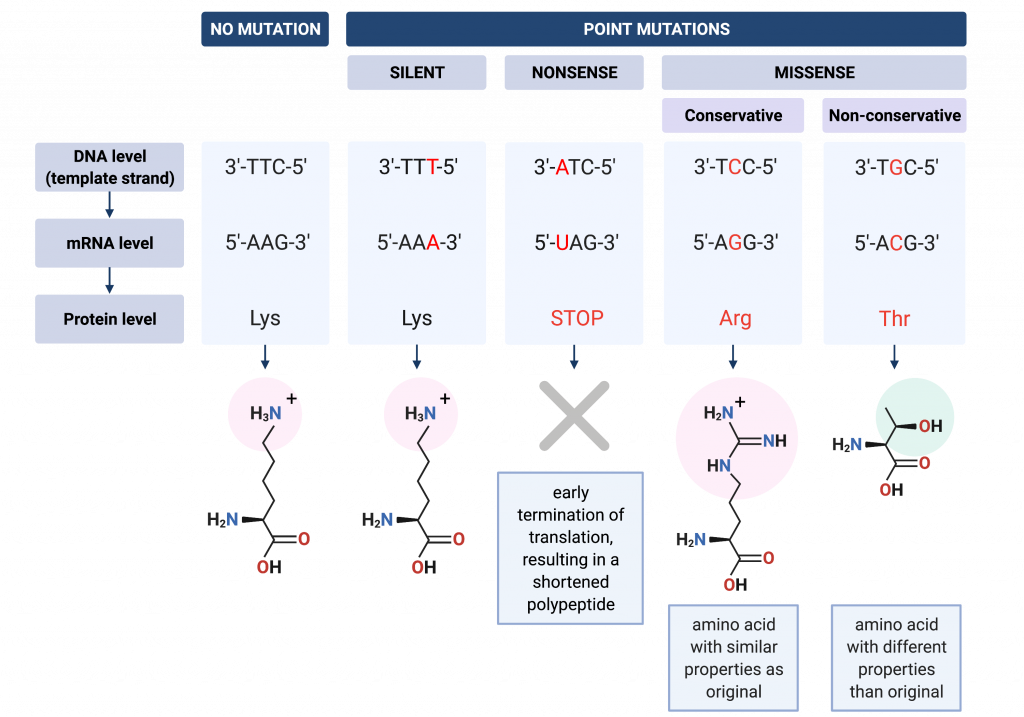
Small-scale mutations refer to changes in one or a few nucleotides. These include insertions, deletions, and substitutions. A substitution mutation, or point mutation, is when a single base pair is changed for a different one. When a point mutation is found within the coding region of a gene, we can further classify it by how it affects the sequence of the resulting protein. A point mutation may be silent, meaning that it changes a codon to another codon that specifies the same amino acid. A point mutation may be a missense mutation, which changes a codon for one amino acid into a codon for a different amino acid. Finally, a point mutation may be a nonsense mutation, meaning it changes a codon that specifies an amino acid into a stop codon, which signals termination of translation and results in a shortened polypeptide product.
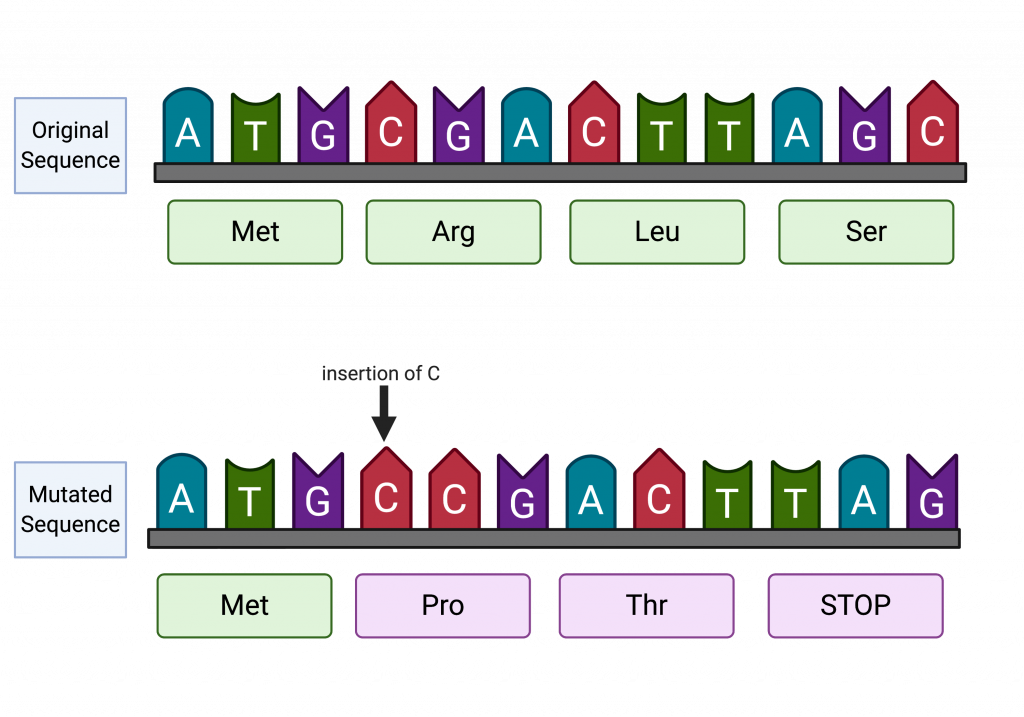
Small-scale insertions and deletions of one or a few bases can have significant effects when they occur within the coding region of a gene. They may change splice sites of a gene, resulting in non-coding regions being included in the mature mRNA or coding regions being absent from the mature mRNA. They can also change the reading frame of a gene, in what is called a frameshift mutation. The resulting protein will probably not be functional because all of the amino acids downstream will be altered. In many cases, frameshift mutations lead to early termination due to a stop codon.
a type of mutation involving the loss of genetic material
a type of mutation that produces an extra copy of one or more nucleotides
a mutation that occurs when a section of DNA breaks and then is reattached in the reverse order
a mutation in which a chromosome breaks and a portion of it is attached to a different nonhomologous chromosome
mutation in which a single base is substituted for a different base
a mutation that changes a codon to a different codon that specifies the same amino acid
a point mutation in which the change results in a codon that specifies a different amino acid than the original codon
a point mutation that results in a stop codon instead of the original codon that specified an amino acid
a small insertion or deletion that changes the reading frame of a gene