12.4 DNA Replication
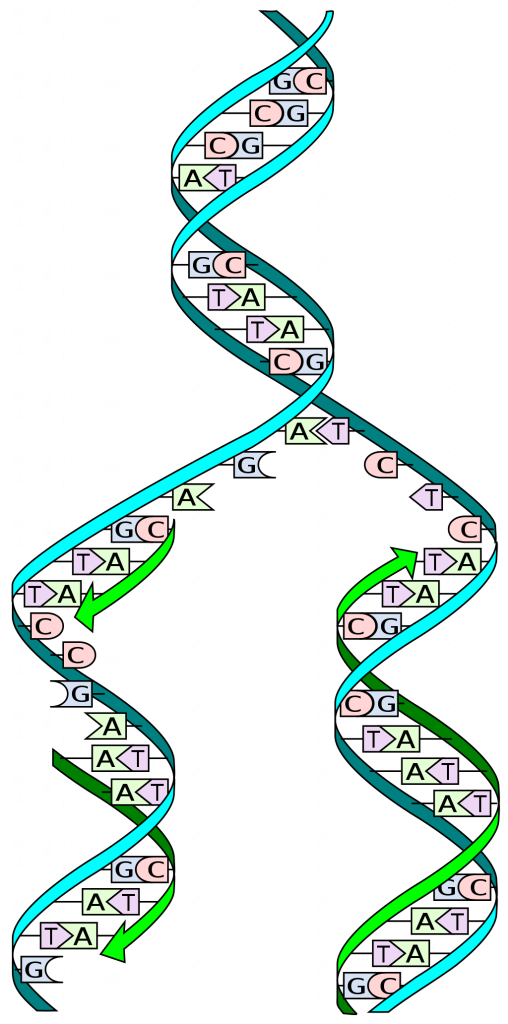
DNA Replication is Semi-Conservative
During DNA replication, each of the two strands that make up the double helix serves as a template from which a new strand is copied. The new strands will be complementary to the parental or “old” strands. When two daughter DNA copies are formed, they have the same sequence and are divided equally into the two daughter cells.
The Process of DNA Replication
DNA replication employs a large number of structural proteins and enzymes, each of which plays a critical role during the process.
How does the replication machinery know where to begin? It turns out that there are specific nucleotide sequences called origins of replication where replication begins. In E. coli, which has a single origin of replication on its one chromosome (as do most prokaryotes), this origin of replication is approximately 245 base pairs long and is rich in AT sequences. The origin of replication is recognized by certain proteins that bind to this site.In eukaryotes, there are multiple origins of replication on each chromosome; for example, humans can have up to 100,000 origins of replication across the genome.
An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required to provide energy for this process. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at each replication origin as the DNA unwinds. The opening of the double helix causes over-winding, or supercoiling, in the double-stranded DNA ahead of the replication fork. These are resolved with the action of topoisomerases.Single-strand binding proteins coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from winding back into a double helix.
Another enzyme, RNA primase, synthesizes an RNA segment that is about five to ten nucleotides long and complementary to the template DNA. Because this sequence primes the DNA synthesis, it is appropriately called the primer. This is necessary because DNA polymerase cannot attach together two DNA mononucleotides. It can only add DNA mononucleotides onto a pre-existing nucleic acid strand. When DNA polymerase extends (polymerizes) a new strand of DNA, it can only add nucleotides to a free 3′-OH group on the growing DNA chain. This means that DNA polymerase can only add nucleotides in the 5′ to 3′ direction.
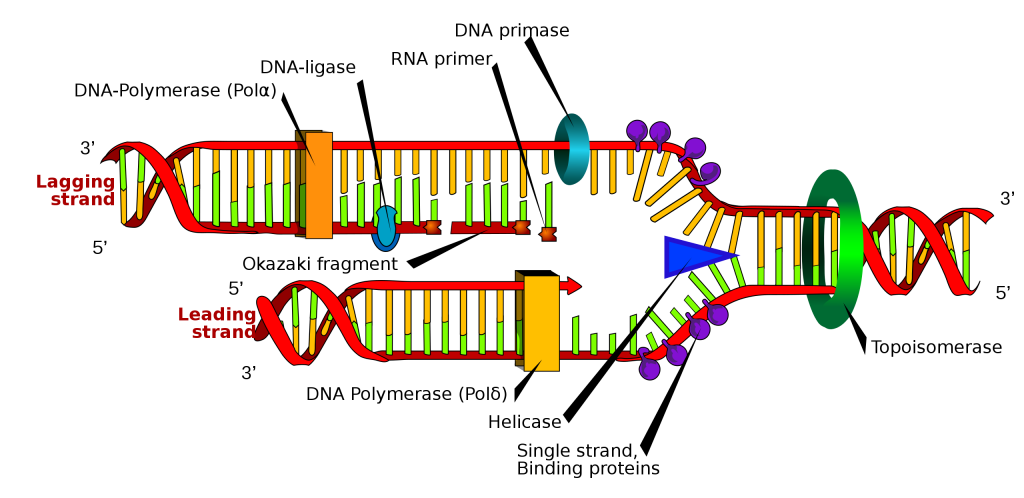
DNA polymerase, also known as DNA pol, adds nucleotides one-by-one to the growing DNA chain that is complementary to the template strand. The addition of nucleotides requires energy; this energy is obtained from the nucleoside triphosphates ATP, GTP, TTP and CTP. Like ATP, the other NTPs (nucleoside triphosphates) are high-energy molecules that can serve both as the source of DNA nucleotides and the source of energy to drive the polymerization. When the bond between the phosphates is “broken,” the energy released is used to form the phosphodiester bond between the incoming nucleotide and the growing chain.
In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III. It is now known that DNA pol III is the enzyme required for DNA synthesis; DNA pol I is an important accessory enzyme in DNA replication, and along with DNA pol II, is primarily required for repair. In eukaryotes, three major DNA polymerases are involved: α, δ and ε. DNA pol α adds a short (20 to 30 nucleotides) DNA fragment to the RNA primer on both strands, and then hands off to a second polymerase. While the leading strand is continuously synthesized by the enzyme pol ε, the lagging strand is synthesized by pol δ.
Because DNA polymerase can only extend in the 5′ to 3′ direction, and because the DNA double helix is antiparallel, there is a challenge or obstacle at the replication fork. Since the two template DNA strands have opposing orientations: only one new DNA strand can be synthesized continuously towards the replication fork. This continuously synthesized strand is known as the leading strand. The other strand is extended away from the replication fork, and is synthesized in numerous small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. New primer segments are laid down in the direction of the replication fork, but each pointing away from it. (Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand.) The leading strand can be extended from a single primer, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3′ to 5′, and that of the leading strand 5′ to 3′.
As synthesis proceeds, the RNA primers are replaced by DNA. In E. coli, the removal of RNA nucleotides and their replacement with DNA nucleotides are both performed by the enzyme DNA polymerase I. The nick that remains between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase, which catalyzes the formation of the phosphodiester linkage between the 3′-OH end of one nucleotide and the 5′ phosphate end of the other fragment.
After the chromosome has been completely replicated, the two DNA copies will be moved into two different cells during cell division.
this enzyme helps to unwind and open up the DNA helix by breaking the hydrogen bonds during replication
Y-shaped structure formed during initiation of replication
enzyme that prevents overwinding of DNA when DNA replication is taking place
enzyme that synthesizes the RNA primer; the primer is needed for DNA polymerase to start synthesis of a new DNA strand
short stretch of nucleotides that is required to initiate replication; in the case of replication, the primer has RNA nucleotides
member of a family of enzymes that catalyzes the addition of new nucleotides to a growing DNA strand
nucleotide triphosphate; a nucleotide with three phosphate groups
strand that is synthesized continuously in the 5'-3' direction, which is synthesized in the direction of the replication fork
DNA fragment that is synthesized in short stretches on the lagging strand
during replication, the strand that is replicated in short fragments and away from the replication fork
enzyme that catalyzes the formation of a phosphodiester linkage between the 3' OH and 5' phosphate ends of the DNA