20.4 Polygenic Inheritance and Epistasis
Mendel’s studies in pea plants implied that the sum of an individual’s phenotype was controlled by genes, such that every characteristic was distinctly and completely controlled by a single gene. However, single observable characteristics are almost always under the influence of multiple genes (each with two or more alleles). For example, multiple genes contribute to eye color in humans.
In some cases, several genes can contribute to aspects of a common phenotype without their gene products ever directly interacting. In the case of organ development, for instance, genes may be expressed sequentially, with each gene adding to the complexity and specificity of the organ. Genes may function in complementary or synergistic fashions, such that two or more genes need to be expressed simultaneously to affect a phenotype. Genes may also oppose each other, with one gene modifying the expression of another.
Epistasis
In epistasis, the interaction between genes is antagonistic, such that one gene masks or interferes with the expression of another. “Epistasis” is a word composed of Greek roots that mean “standing upon.” Often the biochemical basis of epistasis is a gene pathway in which the expression of one gene is dependent on the function of a gene that precedes or follows it in the pathway.
An example of epistasis is pigmentation in mice. The wild-type coat color, agouti (AA), is dominant to solid-colored fur (aa). However, a separate gene (C) is necessary for pigment production. A mouse with a recessive c allele at this locus is unable to produce pigment and is albino regardless of the allele present at locus A. Therefore, the genotypes AAcc, Aacc, and aacc all produce the same albino phenotype. A cross between heterozygotes for both genes (AaCc x AaCc) would generate offspring with a phenotypic ratio of 9 agouti:3 solid color:4 albino. In this case, the C gene is epistatic to the A gene.
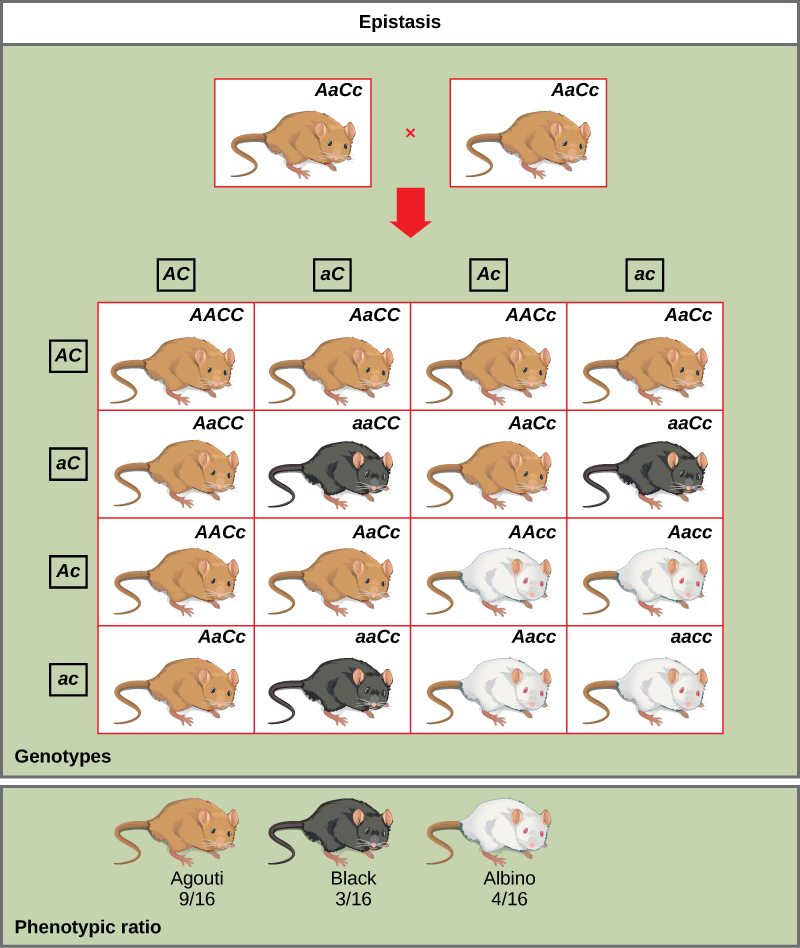
As you work through genetics problems, keep in mind that any single characteristic that results in a phenotypic ratio that totals 16 is typical of a two-gene interaction. Recall the phenotypic inheritance pattern for Mendel’s dihybrid cross, which considered two noninteracting genes—9:3:3:1. Similarly, we would expect interacting gene pairs to also exhibit ratios expressed as 16 parts. Note that we are assuming the interacting genes are not linked; they are still assorting independently into gametes.
Now we will think about this same example of epistasis in a different way – by analyzing a two-step biochemical pathway that represents the mechanism by which fur color is determined in mice:

This diagram suggests that enzyme “C” is able to convert albino fur to black and enzyme “A” is able to convert black fur to agouti. You should recall that every enzyme has an active site that binds to substrates but not to non-substrates. In this example, the active site of enzyme A will bind to the “black molecule” but not to the “albino” molecule. Therefore, a functional enzyme A cannot produce an agouti mouse unless functional enzyme C is also present in the mouse.
Polygenic Inheritance
As the name polygenic inheritance implies, this inheritance mechanism involves multiple genes that work together to determine the phenotype of a particular characteristic (similar to epistasis). However, polygenic inheritance is different from epistasis in that each of these genes encodes the same function. Two examples of polygenic inheritance in humans are height and skin pigmentation.
Let’s imagine that there are 4 different genes that contribute to human height and that two different alleles are possible for each of these genes. One of the alleles for each gene provides a dose of height in an additive way (referred to as an additive allele and represented by a capital letter) whereas the other does not provide such a dose (represented by a lower-case letter and referred to as a non-additive allele). Thus, polygenic inheritance is similar to incomplete dominance except that additional intermediate phenotypes are possible. To simplify the analysis of a polygenic trait, it is assumed that a dose of height contributed by an additive allele of gene A is the same as a dose of height contributed by an additive allele of gene B, etc. Thus, an individual of genotype AaBbCcDd is expected to have the same height as an individual of genotype AAbbccDD since both of these individuals have the same number of additive alleles in their genotypes.
To learn how to recognize polygenic inheritance, we will analyze a cross between two individuals of known genotype: AaBbccDd (3 doses of height) and AaBbccdd (2 doses of height). As with any inheritance mechanism, we must first identify the possible gamete types each parent can produce and place them into a Punnett square. Rather than naming the genotypes of all offspring types from this cross, we will simply count the number of additive alleles.
| ABcD | ABcd | AbcD | aBcD | Abcd | aBcd | abcD | abcd | |
| ABcd | 5 | 4 | 4 | 4 | 3 | 3 | 3 | 2 |
| Abcd | 4 | 3 | 3 | 3 | 2 | 2 | 2 | 1 |
| aBcd | 4 | 3 | 3 | 3 | 2 | 2 | 2 | 1 |
| abcd | 3 | 2 | 2 | 2 | 1 | 1 | 1 | 0 |
Six different phenotypes are expected among the offspring in the ratio: 1/32 with 5 additive alleles, 5/32 with 4 additive alleles, 10/32 with 3 additive alleles, 10/32 with 2 additive alleles, 5/32 with 1 additive allele and 1/32 with 0 additive alleles. Notice that multiple gradations of phenotype are expected, and that offspring with an extreme phenotype are rare (frequency of 0 additive alleles = 1/32) compared to offspring with intermediate phenotypes (frequency of 2 additive alleles = 10/32). Both of these features are expected whenever polygenic inheritance is operative.
antagonistic interaction between genes such that one gene masks or interferes with the expression of another
inheritance of a trait controlled by more than one gene