13.3 Eukaryotic Transcription
Eukaryotes generally have multiple, linear chromosomes. Eukaryotic mRNAs specify a single protein, as opposed to the mRNA transcribed from operons found in prokaryotes. The structure of a generalized eukaryotic protein-coding gene and its product is shown below.
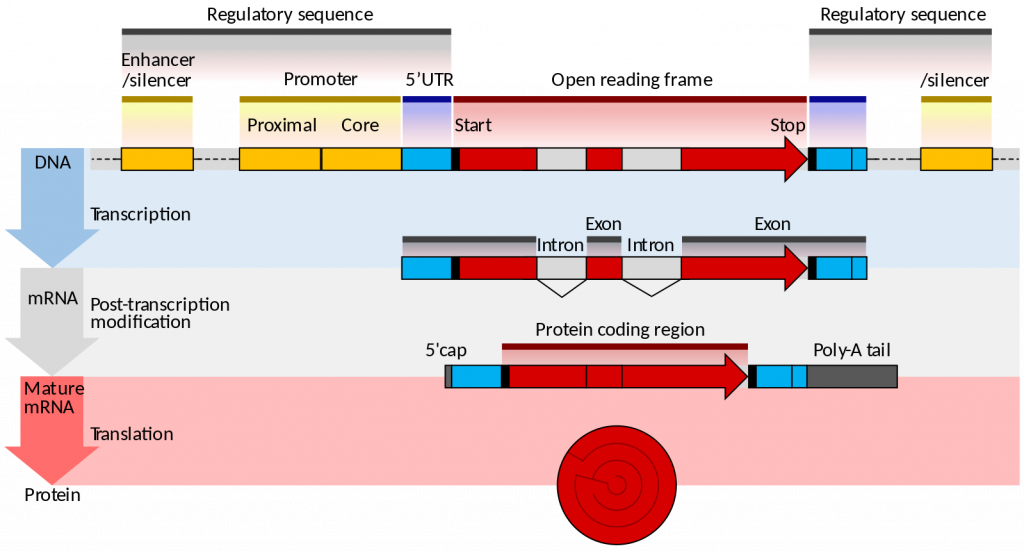
Prokaryotes and eukaryotes perform fundamentally the same process of transcription, with a few key differences. The most important difference between prokaryote and eukaryote transcription is due to the latter’s membrane-bound nucleus and organelles. With the genes bound in a nucleus, the eukaryotic cell must be able to transport its mRNA to the cytoplasm and must protect its mRNA from degrading before it is translated. Eukaryotes also employ three different polymerases that each transcribe a different subset of genes.
| RNA Polymerase | Cellular Compartment | Product of Transcription |
| I | Nucleolus | All rRNAs except 5S rRNA |
| II | Nucleus | All protein-coding nuclear pre-mRNAs and most small nuclear RNAs |
| III | Nucleus | 5S rRNA, tRNAs, and some small nuclear RNAs |
RNA polymerase II is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription but before translation. For clarity, this chapter will use the term “mRNAs” to describe only the mature, processed molecules that are ready to be translated. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes.
RNA Polymerase II Promoters and Transcription Factors
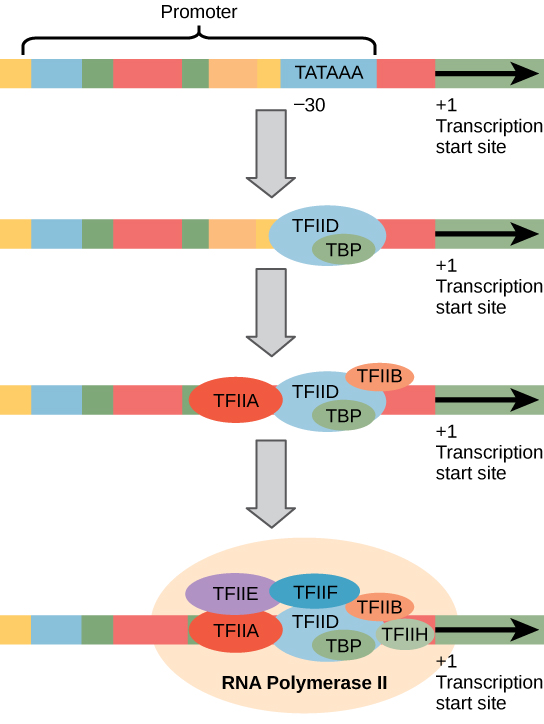
Eukaryotic promoters are much larger and more intricate than prokaryotic promoters. However, both have a sequence similar to the -10 sequence of prokaryotes. In eukaryotes, this sequence is called the TATA box, and has the consensus sequence TATAAA on the coding strand. It is located at -25 to -35 bases relative to the initiation (+1) site.
Like prokaryotic cells, the transcription of genes in eukaryotes requires the action of an RNA polymerase to bind to a DNA sequence upstream of a gene in order to initiate transcription of that gene. However, unlike prokaryotic cells, the eukaryotic RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. There are two types of transcription factors that regulate eukaryotic transcription: General (or basal) transcription factors bind to the core promoter region to assist with the binding of RNA polymerase. Specific transcription factors bind to various regions outside of the core promoter region and interact with the proteins at the core promoter to enhance or repress the activity of the polymerase.
Enhancers and Transcription
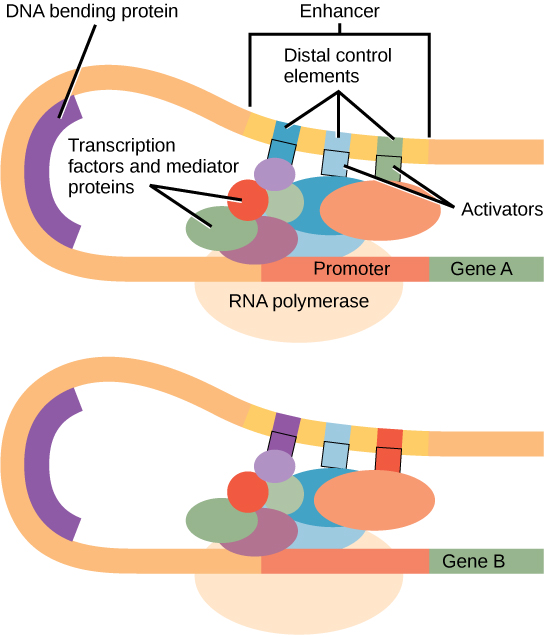
In some eukaryotic genes, there are additional regions that help increase or enhance transcription. These regions, called enhancers or regulatory switches, are not necessarily close to the genes they enhance. They can be located upstream of a gene, within the coding region of the gene, downstream of a gene, and may be thousands of nucleotides away.
Enhancer regions are binding sequences, or sites, for specific transcription factors. When a protein transcription factor binds to its enhancer sequence, the shape of the protein changes, allowing it to interact with proteins at the promoter site. However, since the enhancer region may be distant from the promoter, the DNA must bend to allow the proteins at the two sites to come into physical contact. DNA bending proteins help to bend the DNA and bring the enhancer and promoter regions together. This shape change allows for the interaction of the specific activator proteins bound to the enhancers with the general transcription factors bound to the promoter region and the RNA polymerase.
Eukaryotic Elongation and Termination
Following the formation of the preinitiation complex, the polymerase is released from the promoter, and elongation is allowed to proceed as it does in prokaryotes with the polymerase synthesizing pre-mRNA in the 5′ to 3′ direction. As discussed previously, RNA polymerase II transcribes the major share of eukaryotic genes, so in this section we will focus on how this polymerase accomplishes elongation and termination.
Although the enzymatic process of elongation is essentially the same in eukaryotes and prokaryotes, the DNA template is considerably more complex. When eukaryotic cells are not dividing, their genes exist as a diffuse mass of DNA and proteins called chromatin. The DNA is tightly packaged around charged histone proteins at repeated intervals. These DNA–histone complexes, collectively called nucleosomes, are regularly spaced and include 146 nucleotides of DNA wound around eight histones like thread around a spool.
For polynucleotide synthesis to occur, the transcription machinery needs to move histones out of the way every time it encounters a nucleosome. This is accomplished by a special protein complex called FACT, which stands for “facilitates chromatin transcription.” This complex pulls histones away from the DNA template as the polymerase moves along it. Once the pre-mRNA is synthesized, the FACT complex replaces the histones to recreate the nucleosomes.
The termination of transcription is different for the different polymerases. Unlike in prokaryotes, elongation by RNA polymerase II in eukaryotes takes place 1,000 to 2,000 nucleotides beyond the end of the gene being transcribed. This pre-mRNA tail is subsequently removed by cleavage during mRNA processing. On the other hand, RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves an mRNA hairpin similar to rho-independent termination of transcription in prokaryotes.
nucleotide from which mRNA synthesis proceeds in the 5' to 3' direction; denoted with a “+1”
a protein that binds to DNA and regulates gene expression by either promoting or suppressing transcription
segment of DNA that influences the transcription of a specific gene. can be upstream, downstream, perhaps thousands of nucleotides away, or on another chromosome