8.3 Enzyme Activity
Environmental Influences on Enzyme Activity
The fact that active sites are so perfectly suited to provide specific environmental conditions also means that they are subject to local environmental influences. Increasing the environmental temperature generally increases reaction rates, enzyme-catalyzed or otherwise. However, increasing or decreasing the temperature outside of an optimal range can affect chemical bonds within the active site in such a way that they are less well suited to bind substrates. High temperatures will eventually cause enzymes, like other biological molecules, to denature, a process that changes the substance’s natural properties.
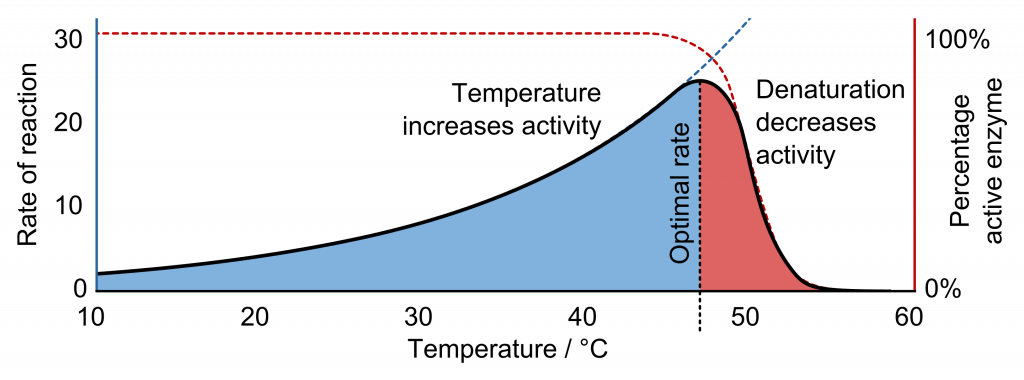
Likewise, the local environment’s pH can also affect enzyme function. Active site amino acid residues have their own acidic or basic properties that are optimal for catalysis. These residues are sensitive to changes in pH that can impair the way substrate molecules bind. Enzymes are suited to function best within a certain pH range, and, as with temperature, extreme environmental pH values (acidic or basic) can cause enzymes to denature.
Enzyme Inhibition
Enzymes can be regulated in ways that either promote or reduce their activity. There are many different kinds of molecules that inhibit or promote enzyme function, and various mechanisms exist for doing so. For example, in some cases of enzyme inhibition, an inhibitor molecule is similar enough to a substrate that it can bind to the active site and simply block the substrate from binding. When this happens, the enzyme is inhibited through competitive inhibition, because an inhibitor molecule competes with the substrate for active site binding.
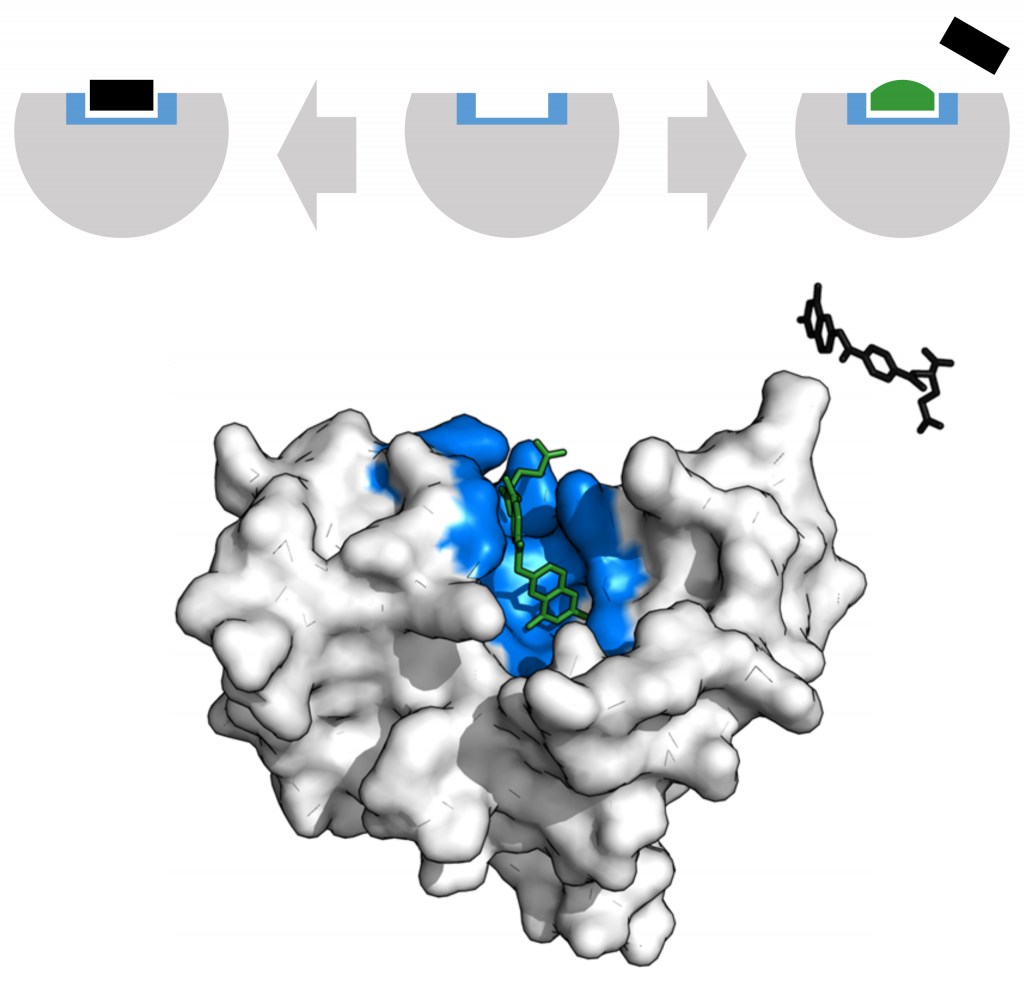
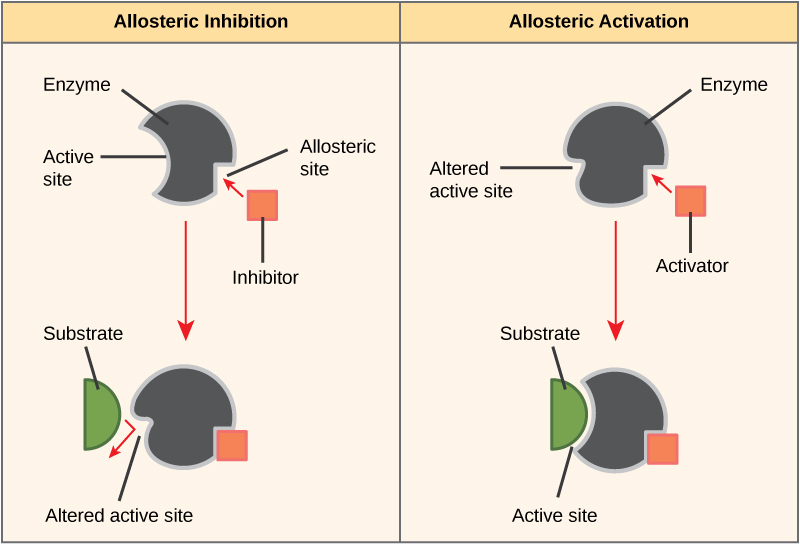
Alternatively, in noncompetitive inhibition, an inhibitor molecule binds to the enzyme at an allosteric site, a binding site away from the active site, and still manages to block substrate binding to the active site.
Some inhibitor molecules bind to enzymes in a location where their binding induces a conformational change that reduces the enzyme’s affinity for its substrate. This type of inhibition is an allosteric inhibition. Most allosterically regulated enzymes have more than one protein subunit. When an allosteric inhibitor binds to an enzyme, all active sites on the protein subunits change slightly such that they bind their substrates with less efficiency. There are allosteric activators as well as inhibitors. Allosteric activators bind to locations on an enzyme away from the active site, inducing a conformational change that increases the affinity of the enzyme’s active site(s) for its substrate(s).
Cofactors and Coenzymes
Many enzymes don’t work optimally, or even at all, unless bound to other specific non-protein helper molecules, either temporarily through ionic or hydrogen bonds or permanently through stronger covalent bonds. Two types of helper molecules are cofactors and coenzymes. Binding to these molecules promotes optimal conformation and function for their respective enzymes. Cofactors are inorganic ions such as iron (Fe++) and magnesium (Mg++). One example of an enzyme that requires a metal ion as a cofactor is the enzyme that builds DNA molecules, DNA polymerase, which requires a bound magnesium ion (Mg++) to function.
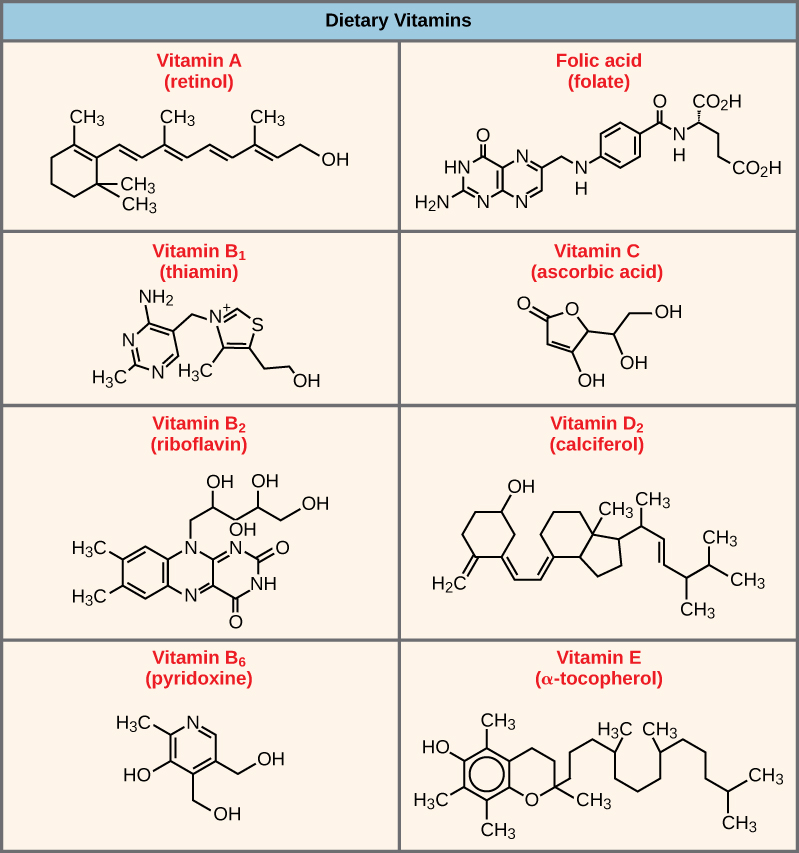
Coenzymes are organic molecules, with a basic atomic structure comprised of carbon and hydrogen, which are required for enzyme action. The most common sources of coenzymes are dietary vitamins. Some vitamins are precursors to coenzymes and others act directly as coenzymes. For example, vitamin C is a coenzyme for multiple enzymes that take part in building the important connective tissue component, collagen. Many enzymes need both inorganic cofactors and organic coenzymes to function.
Enzyme Compartmentalization
In eukaryotic cells, molecules such as enzymes are usually compartmentalized into different organelles. This allows for yet another level of regulation of enzyme activity. Enzymes required only for certain cellular processes are sometimes housed separately along with their substrates, allowing for more efficient chemical reactions. Examples include the enzymes involved in the latter stages of cellular respiration, which take place exclusively in the mitochondria, and the enzymes involved in digesting cellular debris and foreign materials, located within lysosomes.
loss of shape in a protein; can happen as a result of changes in temperature, pH, or chemical exposure
type of inhibition in which the inhibitor competes with the substrate molecule by binding to the active site
inhibition by a binding event at a site different from the active site, which induces a shape change and reduces the enzyme's affinity for its substrate
inorganic ion required for optimal enzyme activity regulation
small organic molecule, such as a vitamin or its derivative, which is required to enhance an enzyme's activity
small structures that exist within cells and carry out cellular functions