14.2 The Protein Synthesis Machinery
The synthesis of proteins consumes more of a cell’s energy than any other metabolic process. In turn, proteins account for more mass than any other component of living organisms (with the exception of water), and proteins perform many different functions in a cell. The process of translation, or protein synthesis, involves the decoding of an mRNA message into a polypeptide product. Amino acids are covalently strung together by interlinking peptide bonds in lengths ranging from approximately 50 to more than 1000 amino acid residues. Each individual amino acid has an amino group (NH2) and a carboxyl group (COOH). Polypeptides are formed when the amino group of one amino acid forms an amide (i.e., peptide) bond with the carboxyl group of another amino acid. This reaction is catalyzed by ribosomes and generates one water molecule (it is a dehydration reaction).
The Protein Synthesis Machinery
In addition to the mRNA template, many molecules and macromolecules contribute to the process of translation. Translation requires the input of an mRNA template, ribosomes, tRNAs, and various enzymatic factors.
Ribosomes
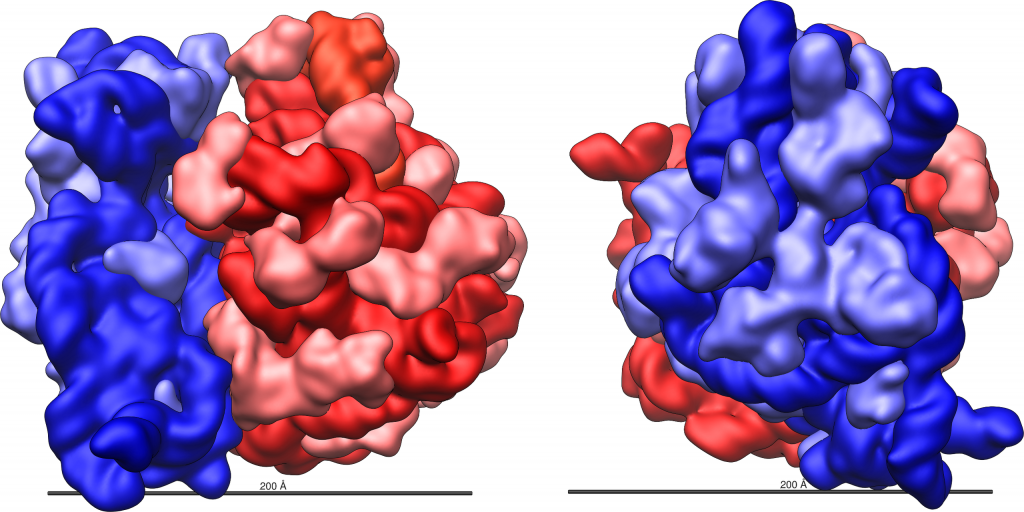
Even before an mRNA is translated, a cell must invest energy to build its ribosomes. In an E. coli cell, there are between 10,000 and 70,000 ribosomes present at any given time. A ribosome is a complex macromolecule composed of ribosomal RNAs (rRNAs), and many polypeptides. In eukaryotes, the nucleolus is completely specialized for the synthesis and assembly of rRNAs.
Ribosomes are found in the cytoplasm of prokaryotes. In eukaryotes, they are also found in the cytoplasm and are sometimes associated with the outer surface of the rough endoplasmic reticulum. Mitochondria and chloroplasts also have their own ribosomes, which look more similar to prokaryotic ribosomes (and have similar drug sensitivities) than the ribosomes just outside their outer membranes in the cytoplasm.
Ribosomes dissociate into large and small subunits when they are not synthesizing proteins and reassociate during the initiation of translation. In E. coli, the small subunit is described as 30S, and the large subunit is 50S, for a total of 70S (Svedberg units are not additive). Mammalian ribosomes have a small 40S subunit and a large 60S subunit, for a total of 80S. The small subunit is responsible for binding the mRNA template, whereas the large subunit sequentially binds tRNAs. Each mRNA molecule is simultaneously translated by many ribosomes (although in different locations along the mRNA), all synthesizing protein in the same direction: reading the mRNA from 5′ to 3′ and synthesizing the polypeptide from the N terminus to the C terminus.
tRNAs
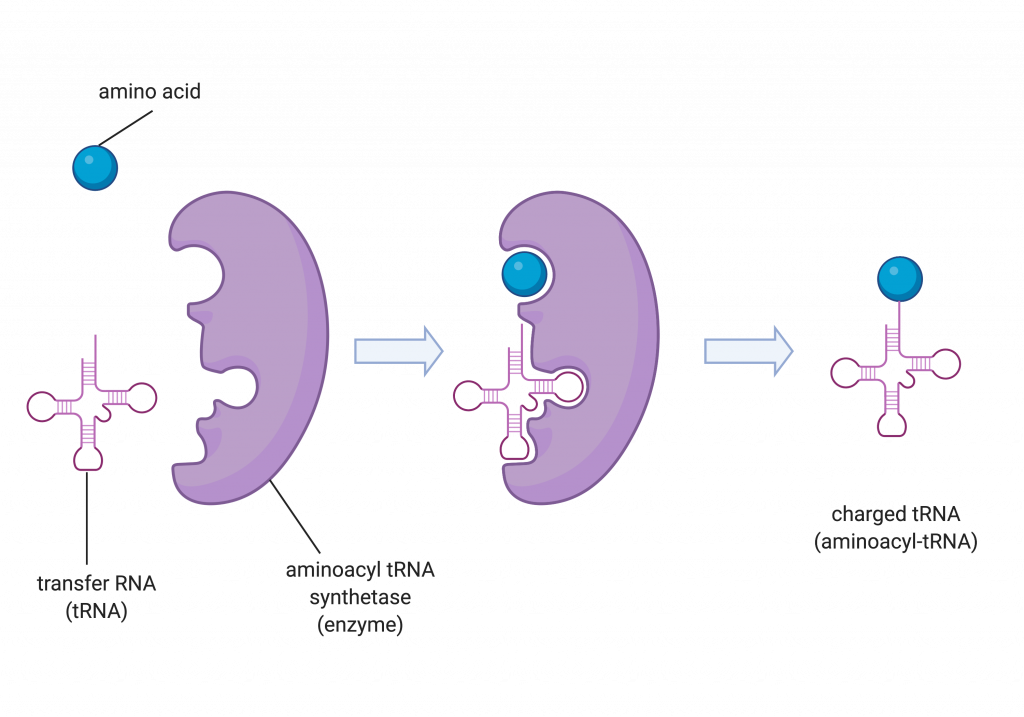
The transfer RNAs (tRNAs) are structural RNA molecules that were transcribed from their corresponding genes by RNA polymerase III. Depending on the species, 40 to 60 types of tRNAs exist in the cytoplasm. Transfer RNAs serve as adaptor molecules. Each tRNA carries a specific amino acid and recognizes one or more of the mRNA codons that define the order of amino acids in a protein. Aminoacyl-tRNAs (tRNA with attached amino acid) bind to the ribosome and add the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins.
Of the 64 possible mRNA codons—or triplet combinations of A, U, G, and C—three specify the termination of protein synthesis and 61 specify the addition of amino acids to the polypeptide chain. Of these 61, one codon (AUG) also encodes the initiation of translation. Each tRNA anticodon can base pair with one or more of the mRNA codons for its amino acid.
Aminoacyl tRNA Synthetases
The process of pre-tRNA synthesis by RNA polymerase III only creates the RNA portion of the adaptor molecule. The corresponding amino acid must be added later, once the tRNA is processed and exported to the cytoplasm. Through the process of tRNA “charging,” each tRNA molecule is linked to its correct amino acid by one of a group of enzymes called aminoacyl tRNA synthetases. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids; the exact number of aminoacyl tRNA synthetases varies by species. The term “charging” is appropriate, since the high-energy bond that attaches an amino acid to its tRNA is later used to drive the formation of the peptide bond. Each tRNA is named for its amino acid.
a protein's monomer; has a central carbon which is attached to an amino group, a carboxyl group, a hydrogen, and an R group or side chain; the R group is different for all 20 common amino acids
a functional group that consists of a nitrogen atom covalently bound to two hydrogen atoms (-NH2)
a functional group that consists of a carbon atom with a double covalent bond to an oxygen atom and a single covalent bond to an oxygen atom that has a single covalent bond to a hydrogen atom (-COOH)
long chain of amino acids linked by peptide (covalent) bonds
cellular structure that synthesizes proteins
region of the endoplasmic reticulum that is studded with ribosomes and engages in protein modification and phospholipid synthesis
a short RNA molecule that links mRNA and a newly synthesized polypeptide by carrying an amino acid to the ribosome
a tRNA that is carrying an amino acid; also called a charged tRNA
enzyme that “charges” tRNA molecules by catalyzing a bond between the tRNA and a corresponding amino acid