23 Lab Protocol: Genetics
Investigating the importance of a large data set using a pair of coins
Collect data sets to analyze:
- Use the percentages in the hypothesis row in the Data Analysis Table as expected frequencies.
- Determine the difference between observed frequency and expected frequency for each category (2 heads, 1 head/1 tail, and 2 tails) and record the difference as a positive number by subtracting the smaller number from the larger number (difference =30-25). Do this for each of the 7 rows.
- Calculate the total difference for each row by adding up the three differences (2 heads + 1 head/1 tail + 2 tails) and record in the final column of table.
- Compare the total difference between expected and observed frequencies (final column of table) according to the size of the data set. Arrange the differences in order from the smallest to the largest number. Record results in the table in your Student Data Sheet.
Analyzing a Dihybrid Cross
It would require at least a year to grow three sequential generations of corn plants. We will therefore not grow or cross corn plants in this lab, but we will analyze purchased corn cobs produced by someone else. We will analyze two different segregating characteristics: purple versus yellow color, and smooth versus wrinkled kernels. Whenever you are asked to describe phenotypes, choose from the following four possibilities: purple smooth, yellow smooth, purple wrinkled, and yellow wrinkled. Keep in mind that true-breeding individuals will be homozygous for the genes involved.
- What phenotype(s) is/are associated with the two true-breeding parental cobs? Also indicate the frequency of each.
-
- Parent cob 1:
- Parent cob 2:
-
- Can we determine if purple is dominant over yellow or if smooth is dominant over wrinkled by looking at the parental cobs? Explain.
- What phenotype(s) is/are associated with the F1 cob? Also indicate the frequency of each phenotype:
- Which is the dominant phenotype?
-
-
- Purple or yellow?
- Smooth or wrinkled?
-
-
- Write a legend to assign names to the genes and the alleles associated with the corn kernels.
- Indicate the genotypes of both parents and of the F1 generation.
- Indicate the genotypes of another set of true-breeding parents that could also produce dihybrid F1 offspring.
- Draw a Punnett square describing a cross between two F1 individuals. Be sure to display the gamete types produced by each parent and the genotypes of the F2 offspring in their proper locations.
- Add offspring phenotypes to the Punnett square.
- Based on the Punnett square, fill in the second and third columns of the data table in your Student Data Sheet.
- Count the kernels of the F2 generation cobs according to their phenotypes and place in the 4th column of the data table. Randomly count at least 100 kernels.
12. Calculate the frequency of each kernel type as a percentage and place into 5th column.
13. Speculate why the percentages in column 3 might not be the same as in column 5.
Using a Pedigree chart to predict the likelihood that a particular child will be colorblind, an X-linked recessive trait
Analyze the following pedigree chart that displays the inheritance of colorblindness.
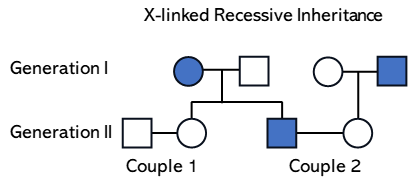
- Write a legend to describe the allele symbols you will use.
- Indicate all that you can about the genotype of each individual without analyzing any relationships between individuals.
- Now look at relationships to complete each genotype. List both genotypes if two different genotypes are possible for any individual.
- Use a Punnett square to predict the probability of colorblindness.
-
-
- First daughter of couple 1:
- First son of couple 1:
- First daughter of couple 2:
- First son of couple 2:
-
-
Analysis of human blood type
Sometimes a geneticist is asked to use the phenotypes of family members to predict their genotypes. One example of this is using the phenotypes of children to determine the genotypes of the parents. A good way to do this is to construct a Punnett square placing the phenotypes of the children in the offspring squares and working backwards to predict gamete types for each parent. These gamete types can then be used to determine the genotype of each parent.
Story Problems:
Story problem 1: Father has type A+ blood and mother has type B- blood. The couple has six children with the following phenotypes: AB+, A-, AB+, A+, A+, AB+. What are the genotypes of the parents assuming that additional children would not introduce other blood types.
Story problem 2: In a paternity case, the mother has blood type O-. What are the possible phenotypes of the father if the child has blood type A+.
Carrying Out a Blood Typing Experiment
Design and carry out an experiment to determine the blood types of 3 different individuals. Use controls of known blood types as you plan and carry out your analysis. Photograph the results of your experiment.
These kits provide 4 blood samples labeled Mr. Green, Ms. Brown, Mr. Smith, and Mr. Jones plus the anti-A, anti-B, and anti-D antisera. It would be good to label the blood samples differently so that we have multiple unknown samples rather than every group doing the same test.